
By Bruna Paulsen
Autism spectrum disorder has been recently associated with hundreds of different risk genes, yet it is unclear whether mutations in these genes converge on a similar neurodevelopmental abnormality. In the Arlotta Lab, we decided to investigate whether three autism-risk genes, SUV420H1, ARID1B, and CHD8, all with very broad hypothetical function, would converge on similar cellular phenotypes.
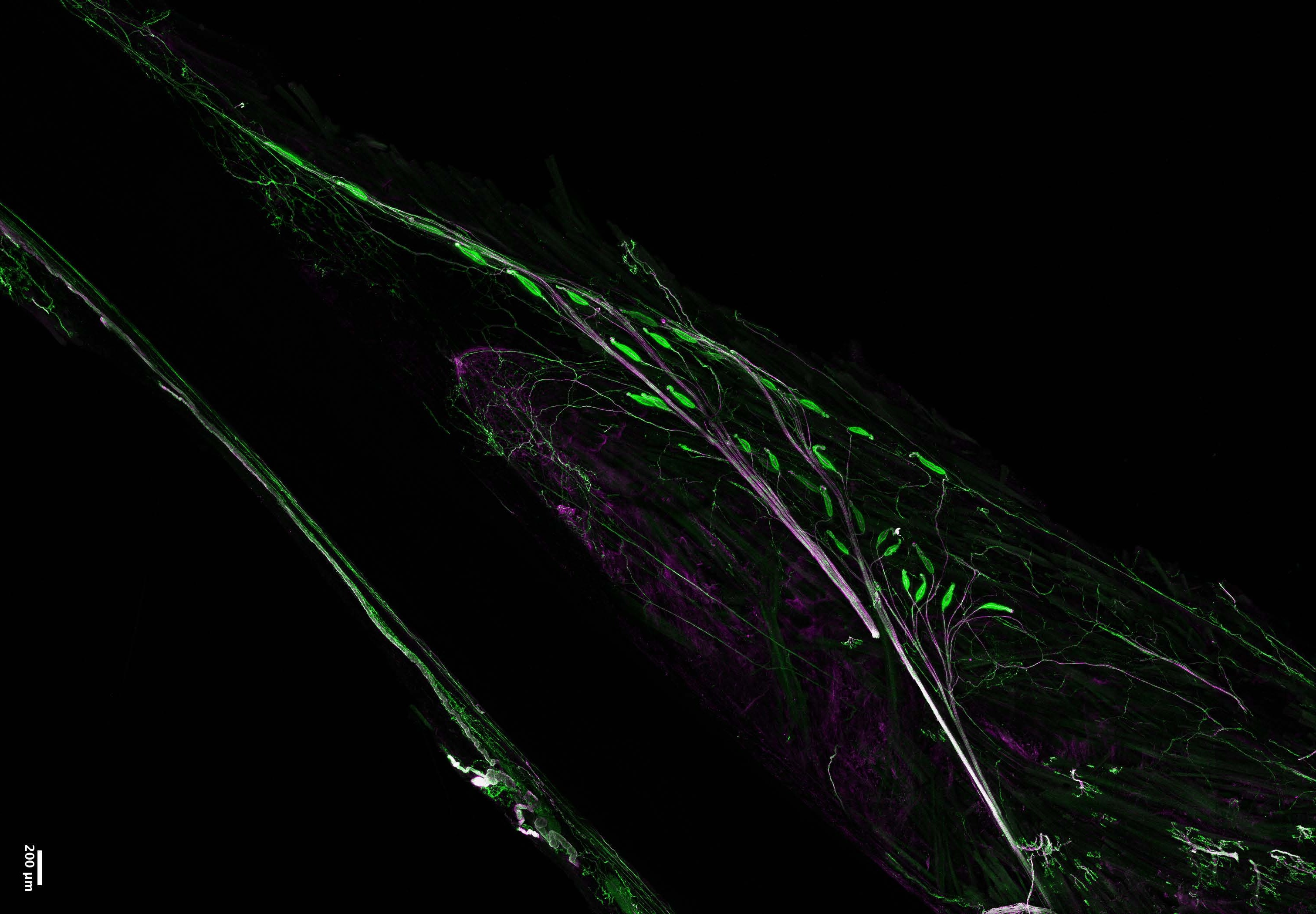
For that, we grew human brain organoids carrying mutations in each of these genes over the course of several months. We found that all the risk genes lead to asynchronous development of specific neurons, although each one acted through unique underlying molecular mechanisms. More specifically, among all the cells generated in these organoids, just the GABAergic neurons and the deep-layer projection neurons were consistently affected, pointing at selected cells that may be special targets in this disorder.

Moreover, we also generated organoids using stem cells from different donor individuals to test how changes in the organoids might be impacted by an individual’s unique genetic background. We observed consistent phenotypic abnormalities across cell lines; however, the level of severity varied across individuals. This suggested that the risk genes’ effects were fine-tuned by the rest of the donor genome.
Our findings that different autism-risk genes converge on a phenotype of asynchronous neuronal development but mostly diverge at the level of molecular targets suggests that a shared clinical pathology of these genes may derive from higher-order processes of neuronal differentiation and circuit wiring. These results encourage future investigation of therapeutic approaches focusing on the modulation of shared dysfunctional circuit properties in addition to shared molecular pathways.
Bruna Paulsen, PhD, is a postdoctoral researcher working in the lab of Paola Arlotta and one of the co-lead authors of this study along with Silvia Velasco, Amanda Kedaigle and Martina Pigoni. The authors thank all of their collaborators, noting this study would not have been possible without the combined expertise of many individuals, working together to tackle a complex problem from different angles.
This story also appeared on the Harvard Brain Science Initiative website and the Department of Stem Cell and Regenerative Biology.
Learn more in the original research article:
Autism genes converge on asynchronous development of shared neuron classes. Paulsen B, Velasco S, Kedaigle AJ, Pigoni M, Quadrato G, Deo AJ, Adiconis X, Uzquiano A, Sartore R, Yang SM, Simmons SK, Symvoulidis P, Kim K, Tsafou K, Podury A, Abbate C, Tucewicz A, Smith SN, Albanese A, Barrett L, Sanjana NE, Shi X, Chung K, Lage K, Boyden ES, Regev A, Levin JZ, Arlotta P. Nature. 2022 Feb;602(7896):268-273.